
BIO-HELIX - RK004-0100
RScript™ miRNA cDNA Synthesis Kit (Stem-loop)
Size: 100 rxns
This kit is developed based on RScript II reverse transcriptase, which uses a reverse transcription primer with a stem-loop structure binding to the 3' end of the miRNA molecule. Under the action of RScript II reverse transcriptase, the artificially elongated miRNA first-strand cDNA is obtained. The recommended general stem-loop sequence is 5'-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAC-3'; other stem-loop sequences can also be selected according to experimental needs. Typically, reverse transcription primers only need to add 6 reverse, complementary bases at the 3' end of the miRNA to the stem-loop sequence. This method is highly specific and only conducts reverse transcription on the mature miRNAs without being interfered by its precursors. The miRNAs with highly homologous sequences can be accurately distinguished. The reverse transcription products obtained with this product can be directly used for subsequent quantitative detection by dye or probe methods.


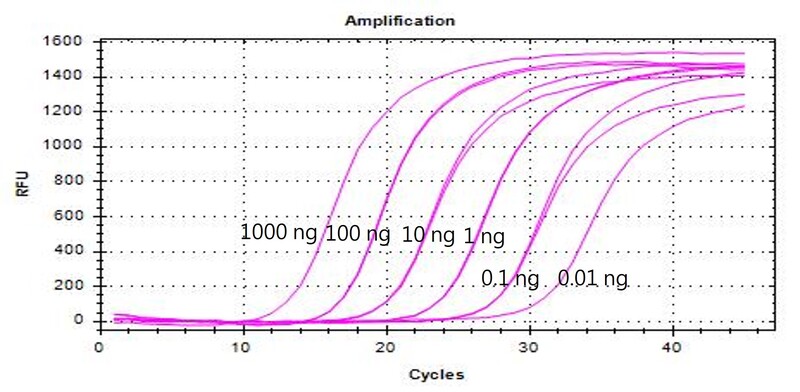
Figure 1. Sensitivity Test of RK004-0050
RK004-0050 was tested with different concentrations (1000 ng, 100 ng, 10 ng, 1 ng, 0.1 ng, 0.01 ng) of Hela cell RNA as templates, targeting the has-mir16-5p gene. Amplification curves demonstrated a sensitivity as low as 10 pg total RNA
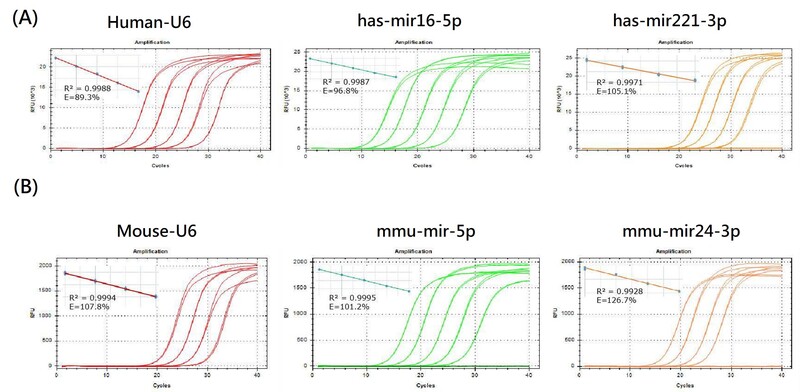
Figure 2. Amplification Performance of RK004-0050 on Templates from Different Sources (A) Hela cell RNA (B) mouse RNA
RK004-0050 was used with different concentrations (1000 ng, 100 ng, 10 ng, 1 ng, 0.1 ng) of Hela cell RNA and mouse RNA as templates. Amplification curves for various miRNAs showed excellent performance for Human-U6, has-mir16-5p, has-mir221-3p, Mouse-U6, mmu-mir-5p, mmu-mir24-3p, among others.
| Name | Download |
|---|---|
| PROTOCOL | Bio-Helix_RK004-0100_Protocol_V3_20240425.pdf |
| Safety Data Sheet|SDS | Bio-Helix_RK004-0050_MSDS_V1.pdf |